The NMODL Framework is a code generation engine for NEURON MODeling Language (NMODL). It is designed with modern compiler and code generation techniques to:
- Provide modular tools for parsing, analysing and transforming NMODL
- Provide easy to use, high level Python API
- Generate optimised code for modern compute architectures including CPUs, GPUs
- Flexibility to implement new simulator backends
- Support for full NMODL specification
Simulators like NEURON use NMODL as a domain specific language (DSL) to describe a wide range of membrane and intracellular submodels. Here is an example of exponential synapse specified in NMODL:
NEURON {
POINT_PROCESS ExpSyn
RANGE tau, e, i
NONSPECIFIC_CURRENT i
}
UNITS {
(nA) = (nanoamp)
(mV) = (millivolt)
(uS) = (microsiemens)
}
PARAMETER {
tau = 0.1 (ms) <1e-9,1e9>
e = 0 (mV)
}
ASSIGNED {
v (mV)
i (nA)
}
STATE {
g (uS)
}
INITIAL {
g = 0
}
BREAKPOINT {
SOLVE state METHOD cnexp
i = g*(v - e)
}
DERIVATIVE state {
g' = -g/tau
}
NET_RECEIVE(weight (uS)) {
g = g + weight
}See INSTALL.md for detailed instructions to build the NMODL from source.
To quickly test the NMODL Framework's analysis capabilities we provide a docker image, which includes the NMODL Framework python library and a fully functional Jupyter notebook environment. After installing docker and docker-compose you can pull and run the NMODL image from your terminal.
To try Python interface directly from CLI, you can run docker image as:
docker run -it --entrypoint=/bin/sh bluebrain/nmodl
And try NMODL Python API discussed later in this README as:
$ python3
Python 3.6.8 (default, Apr 8 2019, 18:17:52)
>>> from nmodl import dsl
>>> import os
>>> examples = dsl.list_examples()
>>> nmodl_string = dsl.load_example(examples[-1])
...
To try Jupyter notebooks you can download docker compose file and run it as:
wget "https://raw.githubusercontent.com/BlueBrain/nmodl/master/docker/docker-compose.yml"
DUID=$(id -u) DGID=$(id -g) HOSTNAME=$(hostname) docker-compose upIf all goes well you should see at the end status messages similar to these:
[I 09:49:53.923 NotebookApp] The Jupyter Notebook is running at:
[I 09:49:53.923 NotebookApp] http://(4c8edabe52e1 or 127.0.0.1):8888/?token=a7902983bad430a11935
[I 09:49:53.923 NotebookApp] Use Control-C to stop this server and shut down all kernels (twice to skip confirmation).
To access the notebook, open this file in a browser:
file:///root/.local/share/jupyter/runtime/nbserver-1-open.html
Or copy and paste one of these URLs:
http://(4c8edabe52e1 or 127.0.0.1):8888/?token=a7902983bad430a11935
Based on the example above you should then open your browser and navigate to the URL http://127.0.0.1:8888/?token=a7902983bad430a11935.
You can open and run all example notebooks provided in the examples folder. You can also create
new notebooks in my_notebooks, which will be stored in a subfolder notebooks at your current
working directory.
Once the NMODL Framework is installed, you can use the Python parsing API to load NMOD file as:
from nmodl import dsl
examples = dsl.list_examples()
nmodl_string = dsl.load_example(examples[-1])
driver = dsl.NmodlDriver()
modast = driver.parse_string(nmodl_string)The parse_file API returns Abstract Syntax Tree (AST) representation of input NMODL file. One can look at the AST by converting to JSON form as:
>>> print (dsl.to_json(modast))
{
"Program": [
{
"NeuronBlock": [
{
"StatementBlock": [
{
"Suffix": [
{
"Name": [
{
"String": [
{
"name": "POINT_PROCESS"
}
...Every key in the JSON form represent a node in the AST. You can also use visualization API to look at the details of AST as:
from nmodl import ast
ast.view(modast)
which will open AST view in web browser:
The central Program node represents the whole MOD file and each of it's children represent the block in the input NMODL file. Note that this requires X-forwarding if you are using Docker image.
Once the AST is created, one can use exisiting visitors to perform various analysis/optimisations. One can also easily write his own custom visitor using Python Visitor API. See Python API tutorial for details.
NMODL Frameowrk also allows to transform AST representation back to NMODL form as:
>>> print (dsl.to_nmodl(modast))
NEURON {
POINT_PROCESS ExpSyn
RANGE tau, e, i
NONSPECIFIC_CURRENT i
}
UNITS {
(nA) = (nanoamp)
(mV) = (millivolt)
(uS) = (microsiemens)
}
PARAMETER {
tau = 0.1 (ms) <1e-09,1000000000>
e = 0 (mV)
}
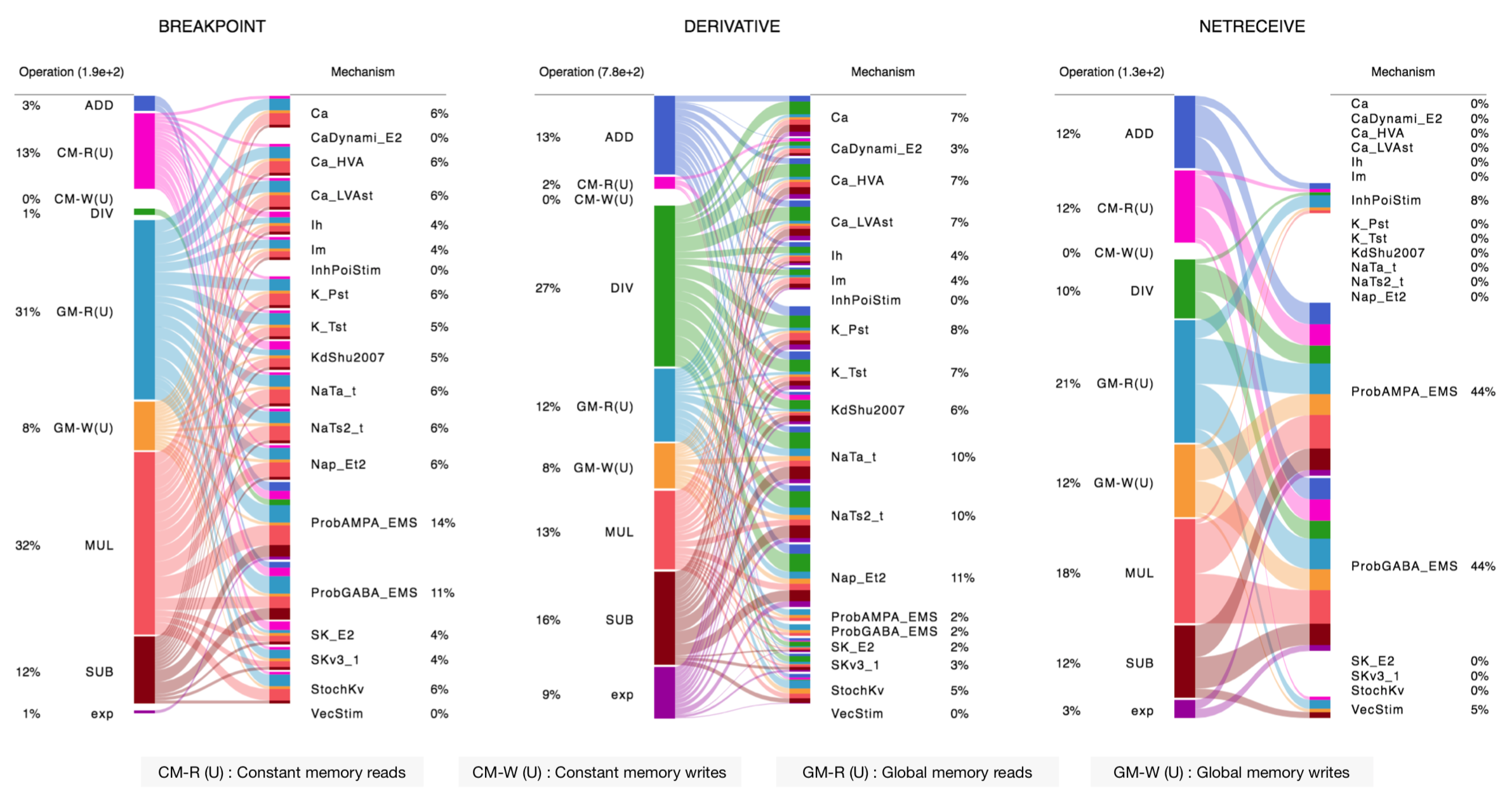
...The NMODL Framework provides rich model introspection and analysis capabilities using various visitors. Here is an example of theoretical performance characterisation of channels and synapses from rat neocortical column microcircuit published in 2015:
To understand how you can write your own introspection and analysis tool, see this tutorial.
Once analysis and optimization passes are performed, the NMODL Framework can generate optimised code for modern compute architectures including CPUs (Intel, AMD, ARM) and GPUs (NVIDIA, AMD) platforms. For example, C++, OpenACC, OpenMP, CUDA and ISPC backends are implemented and one can choose these backends on command line as:
$ nmodl expsyn.mod host --ispc acc --cuda sympy --analytic
Here is an example of generated ISPC kernel for DERIVATIVE block :
export void nrn_state_ExpSyn(uniform ExpSyn_Instance* uniform inst, uniform NrnThread* uniform nt ...) {
uniform int nodecount = ml->nodecount;
const int* uniform node_index = ml->nodeindices;
const double* uniform voltage = nt->actual_v;
int uniform start = 0;
int uniform end = nodecount;
foreach (id = start ... end) {
int node_id = node_index[id];
double v = voltage[node_id];
inst->g[id] = inst->g[id] * vexp( -nt->dt / inst->tau[id]);
}
}To know more about code generation backends, see here. NMODL Framework provides number of options (for code generation, optimization passes and ODE solver) which can be listed as:
$ nmodl -H
NMODL : Source-to-Source Code Generation Framework [version]
Usage: /path/<>/nmodl [OPTIONS] file... [SUBCOMMAND]
Positionals:
file TEXT:FILE ... REQUIRED One or more MOD files to process
Options:
-h,--help Print this help message and exit
-H,--help-all Print this help message including all sub-commands
--verbose=info Verbose logger output (trace, debug, info, warning, error, critical, off)
-o,--output TEXT=. Directory for backend code output
--scratch TEXT=tmp Directory for intermediate code output
--units TEXT=/path/<>/nrnunits.lib
Directory of units lib file
Subcommands:
host
HOST/CPU code backends
Options:
--c C/C++ backend (true)
--omp C/C++ backend with OpenMP (false)
--ispc C/C++ backend with ISPC (false)
acc
Accelerator code backends
Options:
--oacc C/C++ backend with OpenACC (false)
--cuda C/C++ backend with CUDA (false)
sympy
SymPy based analysis and optimizations
Options:
--analytic Solve ODEs using SymPy analytic integration (false)
--pade Pade approximation in SymPy analytic integration (false)
--cse CSE (Common Subexpression Elimination) in SymPy analytic integration (false)
--conductance Add CONDUCTANCE keyword in BREAKPOINT (false)
passes
Analyse/Optimization passes
Options:
--inline Perform inlining at NMODL level (false)
--unroll Perform loop unroll at NMODL level (false)
--const-folding Perform constant folding at NMODL level (false)
--localize Convert RANGE variables to LOCAL (false)
--global-to-range Convert GLOBAL variables to RANGE (false)
--localize-verbatim Convert RANGE variables to LOCAL even if verbatim block exist (false)
--local-rename Rename LOCAL variable if variable of same name exist in global scope (false)
--verbatim-inline Inline even if verbatim block exist (false)
--verbatim-rename Rename variables in verbatim block (true)
--json-ast Write AST to JSON file (false)
--nmodl-ast Write AST to NMODL file (false)
--json-perf Write performance statistics to JSON file (false)
--show-symtab Write symbol table to stdout (false)
codegen
Code generation options
Options:
--layout TEXT:{aos,soa}=soa Memory layout for code generation
--datatype TEXT:{float,double}=soa Data type for floating point variables
--force Force code generation even if there is any incompatibility
--only-check-compatibility Check compatibility and return without generating code
--opt-ionvar-copy Optimize copies of ion variables (false)
We are working on user documentation, you can find current drafts of :
If you would like to know more about the the NMODL Framework, see following paper:
- Pramod Kumbhar, Omar Awile, Liam Keegan, Jorge Alonso, James King, Michael Hines and Felix Schürmann. 2019. An optimizing multi-platform source-to-source compiler framework for the NEURON MODeling Language. In Eprint : arXiv:1905.02241
If you see any issue, feel free to raise a ticket. If you would like to improve this framework, see open issues and contribution guidelines.
The benchmarks used to test the performance and parsing capabilities of NMODL Framework are currently being migrated to GitHub. These benchmarks will be published soon in following repositories:
See contributors.
This work has been funded by the EPFL Blue Brain Project (funded by the Swiss ETH board), NIH grant number R01NS11613 (Yale University) and partially funded by the European Union's Horizon 2020 Framework Programme for Research and Innovation under Grant Agreement number 785907 (Human Brain Project SGA2).