Installation
From PyPI:
pip install cytounet
From source or to use without installing locally:
git clone https://github.com/Nelson-Gon/cytounet.git
# proceed with usual source build procedure
Or:
pip install git+https://github.com/Nelson-Gon/cytounet.gitImport
from cytounet.model import *
from cytounet.data import *
from cytounet.augmentation import *
from cytounet.post_model import *Detailed Sample Usage
Please see the following examples:
Visually:
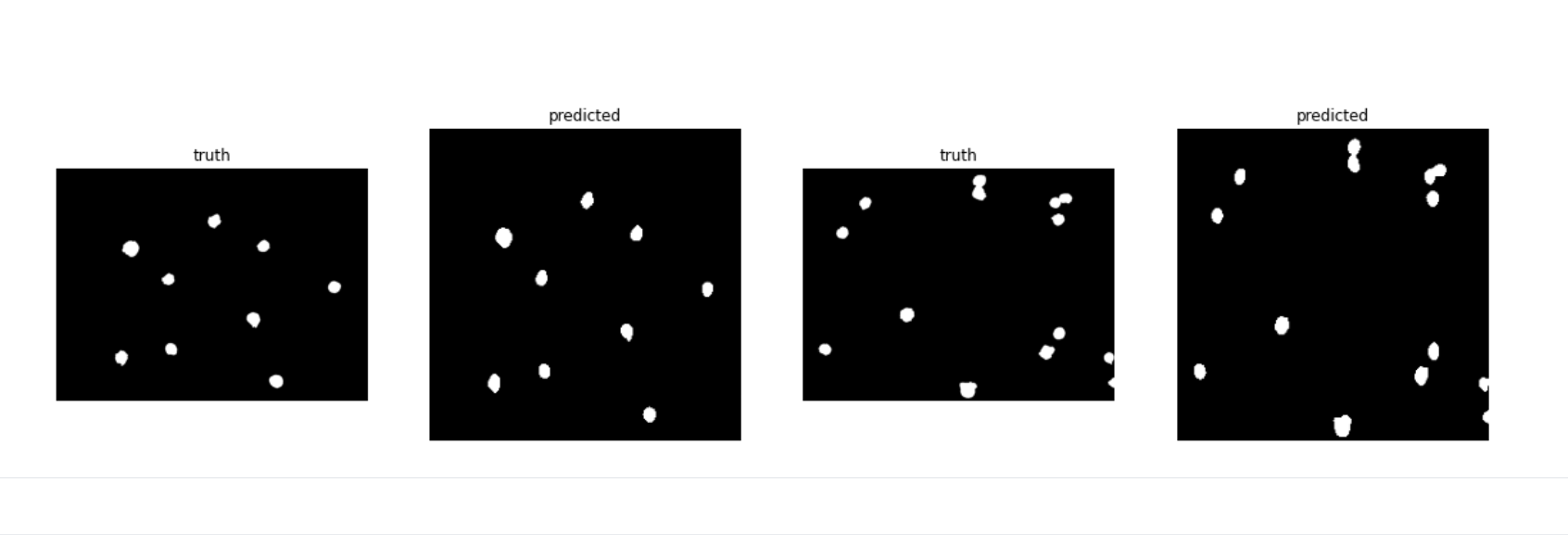
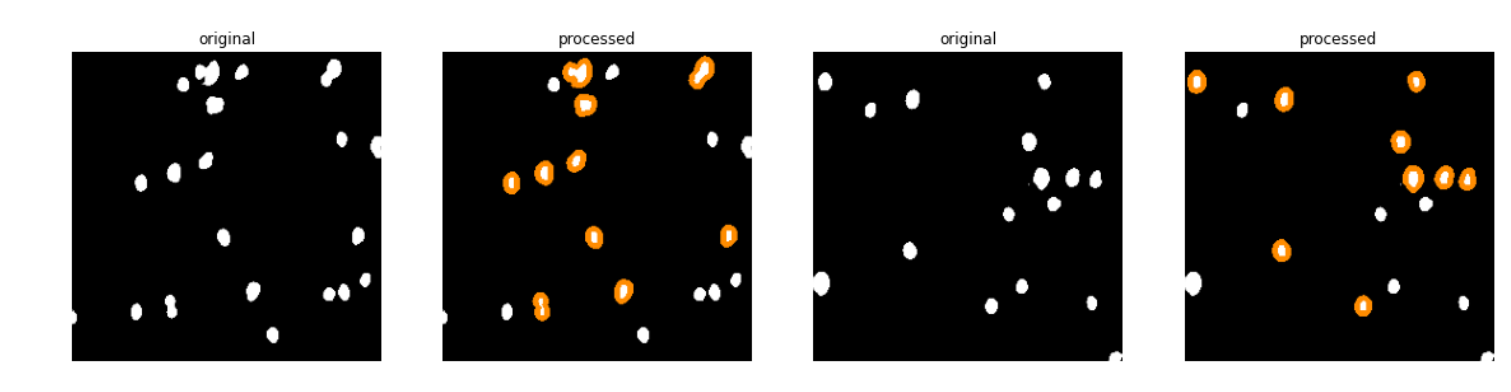
Sample Object Area Visualization(see the typical usage notebook above for detailed usage)
For more examples or to add your own, please see the examples folder.
Is it supported?
A checked box indicates support. You can either add more feature requests here or tackle unchecked boxes and make a pull request to add such support.
-
Single class segmentation
-
Grayscale images
-
Model Validation
-
Determining areas of objects in an image
-
Custom Number of Layers
-
Multi-class segmentation
-
Colored image input
-
COCO Datasets
-
CSV Based Annotations
-
XML Based Annotations
Frequently Asked Questions
Please read our Wiki Pages
To raise an issue or question
If the wiki does not answer your question, please raise a new issue. You can also open an issue if you have any discussion, criticism, or bug reports.
Thank you very much.
References
-
Mouse Embryos Dataset obtained from Broad Bioimage Benchmark Collection. Source.
-
Red Blood Cell Images provided by Anne Carpenter and Roger Wiegand, available here
Credits
- This repository started out as a clone of zhixuhao's original unet implementation.
This repository has considerably diverged from the original implementation hence the need to distribute it separately.
This decision was taken in part due to the relative inactivity of the original implementation which would have made it harder to collaborate. Please take a look at the list of changes from the original implementation.
- The Unet algorithm was introduced by Ronneberger et al. in their paper.