Repository for A Multi-organ Nucleus Segmentation Challenge (MoNuSeg).
Note: If you're interested in using it, feel free to ⭐️ the repo so we know!
- Config File
- Training Graphs
- Patch-Wise Input
- Updation of README Files
- Inference Files
- Quantitative Results
- Visualization of Results
- Train File
- Directory Structure
- Weights Save With Model
Legend
- Resolved
- Work In-Progess
The dataset for this challenge was obtained by carefully annotating tissue images of several patients with tumors of different organs and who were diagnosed at multiple hospitals. This dataset was created by downloading H&E stained tissue images captured at 40x magnification from TCGA archive. H&E staining is a routine protocol to enhance the contrast of a tissue section and is commonly used for tumor assessment (grading, staging, etc.). Given the diversity of nuclei appearances across multiple organs and patients, and the richness of staining protocols adopted at multiple hospitals, the training datatset will enable the development of robust and generalizable nuclei segmentation techniques that will work right out of the box.
Training data containing 30 images and around 22,000 nuclear boundary annotations has been released to the public previously as a dataset article in IEEE Transactions on Medical imaging in 2017.
Test set images with additional 7000 nuclear boundary annotations are available here MoNuSeg 2018 Testing data.
Dataset can be downloaded from Grand Challenge Webiste
A training sample with segmentation mask from training set can be seen below:
| Tissue | Segmentation Mask (Ground Truth) |
|---|---|
 |
 |
Since the size of data set is small and was eaisly loaded into the memmory so we have created patches in online mode. . All the images of training set and test set were reshape to 1024x1024 and then patches were extracted from them. The patch dimensions comprised of 256x256 with 50% overlap among them.
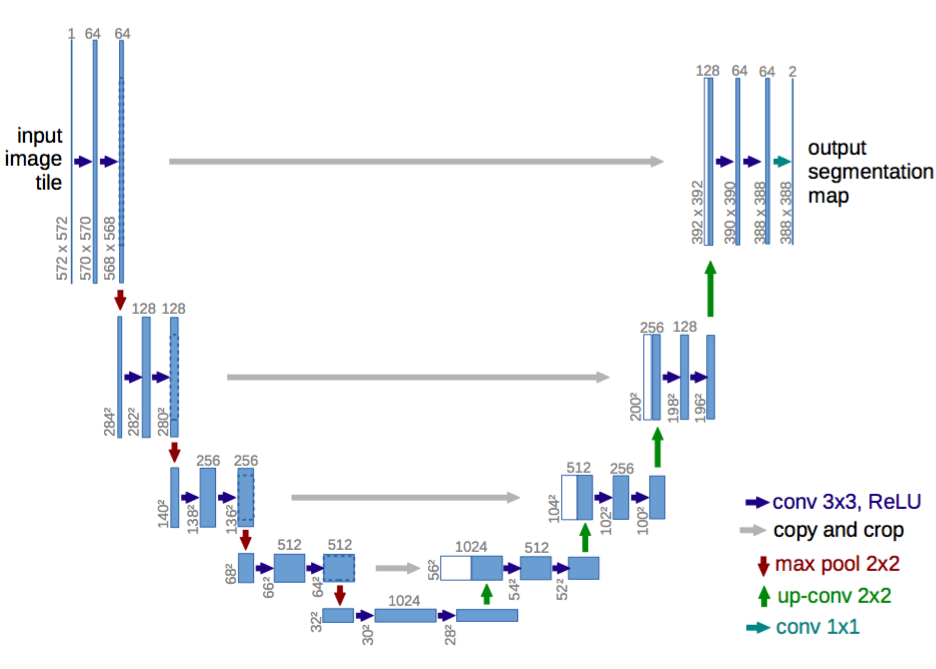
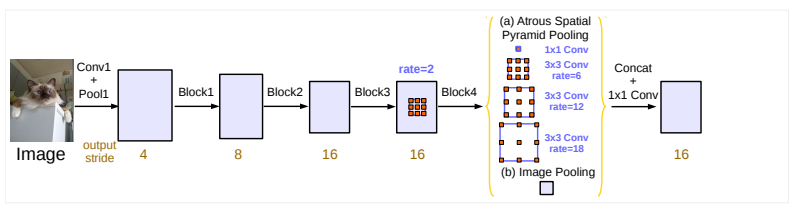
Blocks used to modify U-Net are:
The Pre-Trained models can be downloaded from google drive.
To get this repo work please install all the dependencies using the command below:
pip install -r requirments.txt
To start training run the Train.py script from the command below. For training configurations refer to the config.json file. You can update the file according to your training settings. Model avaible for training are U-NET,SegNet, DeepLabv3+.
python Train.py
To test the trained models on Test Images you first have to download the weights and place them in the results. After downliading the weights you unzip them and then run the Inference by using the command below. For testing configurations please refer to the config.json file.
python Test.py
| Tissue | Mask | Predicted Mask |
|---|---|---|
 |
 |
 |
| Model | Loss | Accuracy | F1 Score | Dice Score |
|---|---|---|---|---|
| Unet | 0.0835 | 0.9150 | 0.7910 | 0.7906 |
| Segnet | 0.4820 | 0.8077 | 0.5798 | 0.3684 |
| DeeplabV3+ | 0.0783 | 0.9120 | 0.7750 | 0.7743 |
| Unet + Skip Connections + ASPP + SE Block | 0.0770 | 0.9210 | 0.801 | 0.8005 |
Three Segmentation models have been trained and the model is evaluated on three metrics namely:
- Accuracy
- F1-Score
- Dice Score
Maintainer Syed Nauyan Rashid ([email protected])