variation graph data structures, interchange formats, alignment, genotyping, and variant calling methods
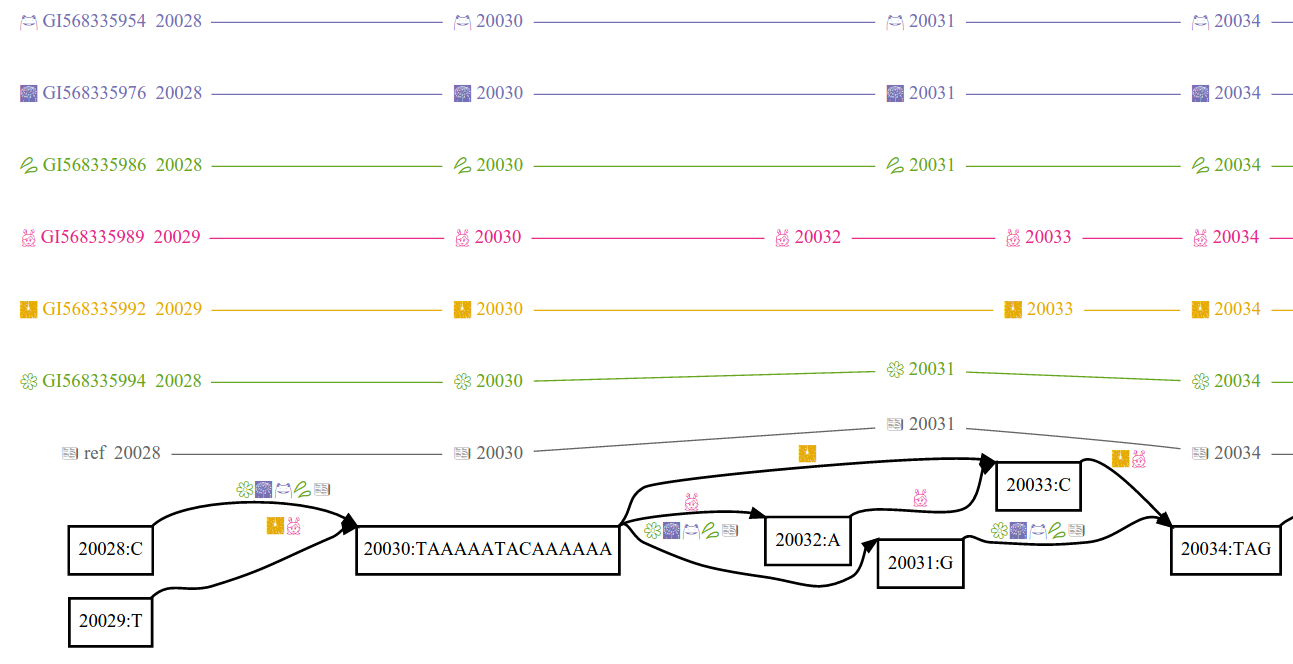
Variation graphs provide a succinct encoding of the sequences of many genomes. A variation graph (in particular as implemented in vg) is composed of:
- nodes, which are labeled by sequences and ids
- edges, which connect two nodes via either of their respective ends
- paths, describe genomes, sequence alignments, and annotations (such as gene models and transcripts) as walks through nodes connected by edges
This model is similar to a number of sequence graphs that have been used in assembly and multiple sequence alignment. Paths provide coordinate systems relative to genomes encoded in the graph, allowing stable mappings to be produced even if the structure of the graph is changed.
The easiest way to get vg is to download one of our release builds for Linux. We have a 6-week release cadence, so our builds are never too far out of date.

Download the latest vg release for Linux
For MacOS, see Building on MacOS.
If you don't want to or can't use a pre-built release of vg, or if you want to become a vg developer, you can build it from source instead.
First, obtain the repo and its submodules:
git clone --recursive https://github.com/vgteam/vg.git
cd vg
Then, install VG's dependencies. You'll need the protobuf and jansson development libraries installed, and to run the tests you will need jq, bc and rs. On Ubuntu, you should be able to do:
make get-deps
On other distros, you will need to perform the equivalent of:
sudo apt-get install build-essential git cmake pkg-config libncurses-dev libbz2-dev \
protobuf-compiler libprotoc-dev libjansson-dev automake libtool \
jq bc rs curl unzip redland-utils librdf-dev bison flex gawk \
lzma-dev liblzma-dev liblz4-dev libffi-dev
At present, you will need GCC version 4.9 or greater to compile vg. (Check your version with gcc --version.)
Other libraries may be required. Please report any build difficulties.
Note that a 64-bit OS is required. Ubuntu 16.04 should work. You will also need a CPU that supports SSE 4.2 to run VG; you can check this with cat /proc/cpuinfo | grep sse4_2.
When you are ready, build with . ./source_me.sh && make, and run with ./bin/vg.
You can also produce a static binary with make static, assuming you have static versions of all the dependencies installed on your system.
The first step is to clone the vg repository:
git clone --recursive https://github.com/vgteam/vg.git
cd vg
VG depends on a number of packages being installed on the system where it is being built. Dependencies can be installed using either MacPorts or Homebrew.
You can use MacPorts to install VG's dependencies:
sudo port install libtool jansson jq cmake pkgconfig autoconf automake libtool coreutils samtools redland bison gperftools md5sha1sum rasqal gmake autogen cairo libomp
Homebrew provides another package management solution for OSX, and may be preferable to some users over MacPorts. VG ships a Brewfile describing its Homebrew dependencies, so from the root vg directory, you can install dependencies, and expose them to vg, like this:
# Install all the dependencies in the Brewfile
brew bundle
# Use GNU versions of coreutils over Apple versions
export PATH="/usr/local/opt/coreutils/libexec/gnubin:/usr/local/bin:$PATH"
# Force use of new version of bison
brew link bison --force
# NOTE! If brew says that it is refusing to link Bison, follow its suggested
# instructions to put Bison on your PATH instead.
# Use glibtool/ize
export LIBTOOL=glibtool
export LIBTOOLIZE=glibtoolize
# Use installed libraries
export LD_LIBRARY_PATH=/usr/local/lib:$LD_LIBRARY_PATH;
export LIBRARY_PATH=$LD_LIBRARY_PATH;
While Apple's clang can build VG, the C++ standard library it uses doesn't support some parallel extensions, so a Clang-built VG will be slower. Better results can be achieved by building with GNU GCC >= 4.9 and its libstdc++ standard library.
With MacPorts, you can install GNU GCC like this:
sudo port install gcc7 clang-3.8
To make GCC 7 the default compiler, run (use none instead of mp-gcc7 to revert back):
sudo port select gcc mp-gcc7
Some OSX users also need to have the MacPorts Clang assembler for building VG's dependencies (use none instead of mp-clang-3.8 to revert back):
sudo port select clang mp-clang-3.8
With Homebrew, you can install GNU GCC for VG like this:
brew install gcc6
# Manually create symlinks to make Homebrew GCC 6 the default gcc and g++
ln -s gcc-6 /usr/local/bin/gcc
ln -s g++-6 /usr/local/bin/g++
With dependencies and compilers installed, VG can now be built:
. ./source_me.sh && make
Note that static binaries cannot yet be built for Mac.
Our team has successfully built vg on Mac with GCC versions 4.9, 5.3, 6, 7, and 7.3, as well as Clang 9.0.
The simplest thing to do with vg is to build a graph and align to it. At present, you'll want to use a reference and VCF file to do so. If you're working in the test/ directory:
vg construct -r small/x.fa -v small/x.vcf.gz >x.vgvg view provides a way to convert the graph into various formats:
# GFA output
vg view x.vg >x.gfa
# dot output suitable for graphviz
vg view -d x.vg >x.dot
# json version of binary alignments
vg view -a x.gam >x.jsonAs this is a small graph, you could align to it using a full-length partial order alignment:
vg align -s CTACTGACAGCAGAAGTTTGCTGTGAAGATTAAATTAGGTGATGCTTG x.vgNote that you don't have to store the graph on disk at all, you can simply pipe it into the local aligner:
vg construct -r small/x.fa -v small/x.vcf.gz | vg align -s CTACTGACAGCAGAAGTTTGCTGTGAAGATTAAATTAGGTGATGCTTG -Most commands allow the streaming of graphs into and out of vg.
If your graph is large, you want to use vg index to store the graph and vg map to align reads. vg map implements a kmer based seed and extend alignment model that is similar to that used in aligners like novoalign or MOSAIK. First an on-disk index is built with vg index which includes the graph itself and kmers of a particular size. When mapping, any kmer size shorter than that used in the index can be employed, and by default the mapper will decrease the kmer size to increase sensitivity when alignment at a particular k fails.
# construct the graph
vg construct -r small/x.fa -v small/x.vcf.gz > x.vg
# store the graph in the xg/gcsa index pair
vg index -x x.xg -g x.gcsa -k 16 x.vg
# align a read to the indexed version of the graph
# note that the graph file is not opened, but x.vg.index is assumed
vg map -s CTACTGACAGCAGAAGTTTGCTGTGAAGATTAAATTAGGTGATGCTTG -x x.xg -g x.gcsa > read.gam
# simulate a bunch of 150bp reads from the graph, one per line
vg sim -n 1000 -l 150 -x x.xg > x.sim.txt
# now map these reads against the graph to get a GAM
vg map -T x.sim.txt -x x.xg -g x.gcsa > aln.gam
# surject the alignments back into the reference space of sequence "x", yielding a BAM file
vg surject -x x.xg -b aln.gam > aln.bam
# or alternatively, surject them to BAM in the call to map
vg sim -n 1000 -l 150 -x x.xg > x.sim.txt
vg map -T x.sim.txt -x x.xg -g x.gcsa --surject-to bam > aln.bamVariation from alignments can be embedded back into the graph. This process is called augmentation and is important for variant calling, for example (see below).
# augment the graph with all variation from the GAM, saving to aug.vg. aug.gam contains the same reads as aln.gam but mapped to aug.vg
vg augment x.vg aln.gam -A aug.gam > aug.vg
# augment the graph with all variation from the GAM, saving each mapping as a path in the graph.
# Note, this can be much less efficient than the above example if there are many alignments in the GAM
vg augment x.vg aln.gam -i > aug_with_paths.vgThe following example shows how to construct a VCF file from a read alignment and graph. Input must be split into chunks (see vg chunk) in order to run on whole genome.
# filter secondary and ambiguous read mappings out of the gam
vg filter alignment.gam -r 0.90 -fu -s 2 -o 0 -D 999 -x graph.xg > filtered.gam
# create an augmented graph by adding variation from the reads
vg augment graph.vg filtered.gam -a pileup -S aug_graph.support -Z aug_graph.trans > aug_graph.vg
# to only recall variants that are already in the graph, add -g 9999999 to the augment options above.
# Make calls by thresholding based on read support for graph path SEQ
vg call aug_graph.vg -b graph.vg -s aug_graph.support -z aug_graph.trans -r SEQ > calls.vcf
# Or Make calls using a Freebayes-like genotyping algorithm for graph path SEQ
vg genotype graph.vg -G alignment.gam -E -v -r SEQ > calls.vcf
# for comparison purposes, it's very useful to normalize the vcf output, especially for more complex graphs which can make large variant blocks that contain a lot of reference bases (Note: requires [vt](http://genome.sph.umich.edu/wiki/Vt)):
vt decompose_blocksub -a calls.vcf | vt normalize -r FASTA_FILE - > calls.clean.vcf
To produce a VCF file for a whole chromosome, the graph must be cut up along the reference genome and called in chunks. scripts/chunked_call wraps this functionality to produce chromosome-sized VCFs in a single command line (from a GAM file and XG index)
A variety of commands are available:
- construct: graph construction
- view: conversion (dot/protobuf/json/GFA)
- index: index features of the graph in a disk-backed key/value store
- find: use an index to find nodes, edges, kmers, or positions
- paths: traverse paths in the graph
- align: local alignment
- map: global alignment (kmer-driven)
- stats: metrics describing graph properties
- join: combine graphs (parallel)
- concat: combine graphs (serial)
- ids: id manipulation
- kmers: generate kmers from a graph
- sim: simulate reads by walking paths in the graph
- mod: various transformations of the graph
- surject: force graph alignments into a linear reference space
- msga: construct a graph from an assembly of multiple sequences
- validate: determine if graph is valid
- filter: filter reads out of an alignment
- augment: adds variation from aligned reads into the graph
- call/genotype: call variants from an augmented graph
vg is a collection of tools based on a common data model (the variation graph) that is described by a protobuf schema (vg.proto). Data objects defined in vg.proto may be serialized via a stream pattern defined in stream.hpp. It is not necessary to write code in vg in order to interface with the algorithms defined here. Rather, it is sometimes simpler to write an external algorithm that reads and writes the same data formats.
MIT