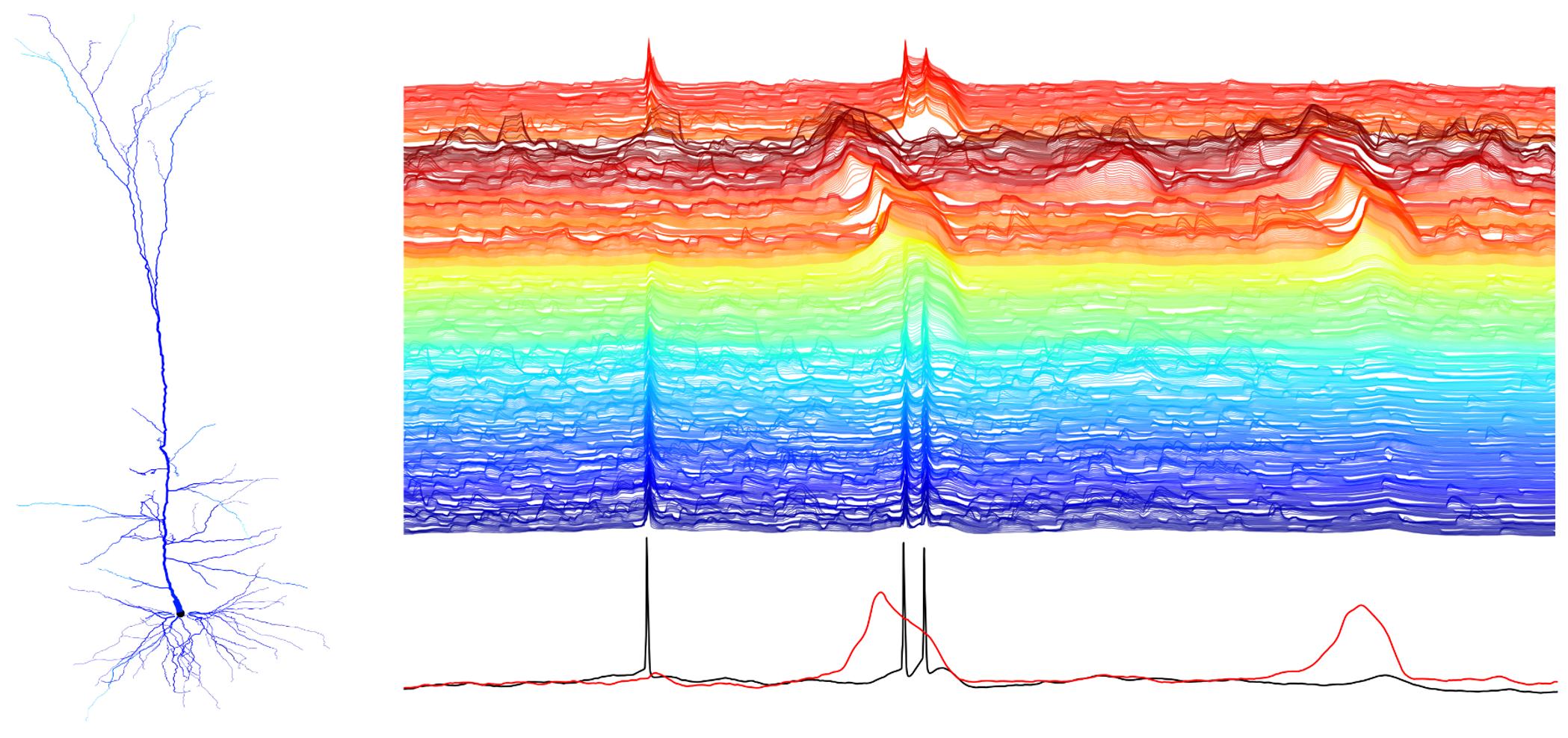
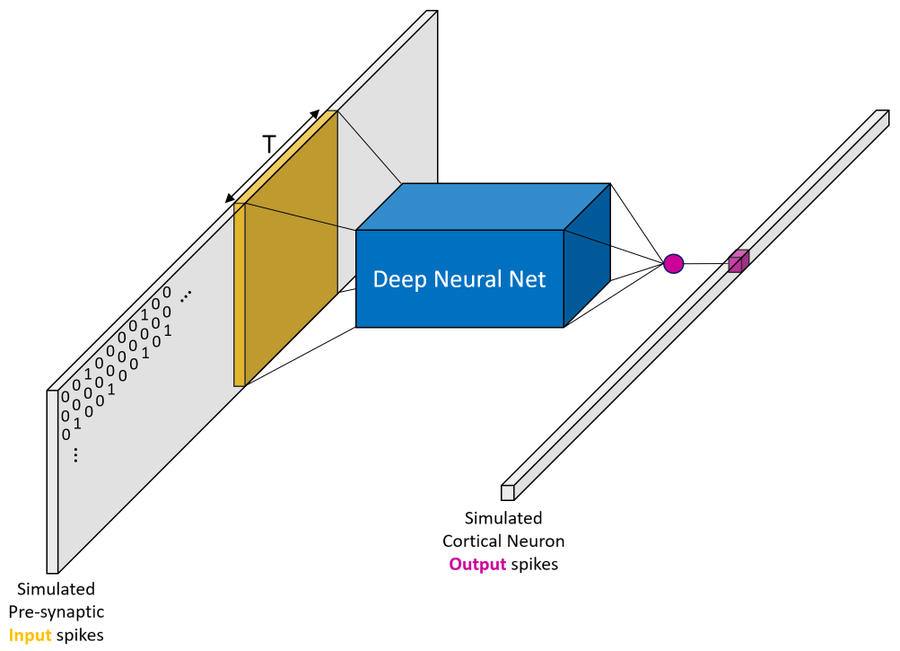
This repo contains the code behind the work "Single Cortical Neurons as Deep Artificial Neural Networks"
David Beniaguev, Idan Segev, Michael London
Abstract: Utilizing recent advances in machine learning, we introduce a systematic approach to characterize neurons’ input/output (I/O) mapping complexity. Deep neural networks (DNNs) were trained to faithfully replicate the I/O function of various biophysical models of cortical neurons at millisecond (spiking) resolution. A temporally convolutional DNN with five to eight layers was required to capture the I/O mapping of a realistic model of a layer 5 cortical pyramidal cell (L5PC). This DNN generalized well when presented with inputs widely outside the training distribution. When NMDA receptors were removed, a much simpler network (fully connected neural network with one hidden layer) was sufficient to fit the model. Analysis of the DNNs’ weight matrices revealed that synaptic integration in dendritic branches could be conceptualized as pattern matching from a set of spatiotemporal templates. This study provides a unified characterization of the computational complexity of single neurons and suggests that cortical networks therefore have a unique architecture, potentially supporting their computational power.
Neuron version of paper: cell.com/neuron/fulltext/S0896-6273(21)00501-8
Open Access (slightly older) version of Paper: biorxiv.org/content/10.1101/613141v2
Dataset and pretrained networks: kaggle.com/selfishgene/single-neurons-as-deep-nets-nmda-test-data
Dataset for training new models: kaggle.com/selfishgene/single-neurons-as-deep-nets-nmda-train-data
Notebook with main result: kaggle.com/selfishgene/single-neuron-as-deep-net-replicating-key-result
Notebook exploring the dataset: kaggle.com/selfishgene/exploring-a-single-cortical-neuron
Twitter thread for short visual summery #1: twitter.com/DavidBeniaguev/status/1131890349578829825
Twitter thread for short visual summery #2: twitter.com/DavidBeniaguev/status/1426172692479287299
Figure360, author presentation of Figure 2 from the paper: youtube.com/watch?v=n2xaUjdX03g
- Use
integrate_and_fire_figure_replication.pyto simulate, fit, evaluate and replicate the introductory figure in the paper (Fig. 1)
- Use
simulate_L5PC_and_create_dataset.pyto simulate a single neuron- All major parameters are documented inside the file using comments
- All necessary NEURON
.hocand.modsimulation files are under the folderL5PC_NEURON_simulation\
- Use
dataset_exploration.pyto explore the generated dataset - Alternativley, just download the data from kaggle, and look at exploration script
- neuron github repo: github.com/neuronsimulator/nrn
- recommended introductory NEURON tutorial: github.com/orena1/NEURON_tutorial
- official NEURON with python tutorial: neuron.yale.edu/neuron/static/docs/neuronpython/index.html
- NEURON help fortum: neuron.yale.edu/phpBB/index.php?sid=31f0839c5c63ca79d80790460542bbf3
fit_CNN.pycontains the code used to fit a network to the datasetevaluate_CNN_test.pyandevaluate_CNN_valid.pycontains the code used to evaluate the performace of the networks on test and validation sets
- Use
main_figure_replication.pyto replicate the main figures (Fig. 2 & Fig. 3) after generating data and training models valid_fitting_results_exploration.pycan be used to explore the fitting results on validation datasettest_accuracy_vs_complexity.pycan be used to generate the model accuracy vs complexity plots (Fig. S5)- Alternativley, visit key figure replication notebook on kaggle
We thank Oren Amsalem, Guy Eyal, Michael Doron, Toviah Moldwin, Yair Deitcher, Eyal Gal and all lab members of the Segev and London Labs for many fruitful discussions and valuable feedback regarding this work.
If you use this code or dataset, please cite the following two works:
- David Beniaguev, Idan Segev and Michael London. "Single cortical neurons as deep artificial neural networks." Neuron. 2021; 109: 2727-2739.e3 doi: https://doi.org/10.1016/j.neuron.2021.07.002
- Hay, Etay, Sean Hill, Felix Schürmann, Henry Markram, and Idan Segev. 2011. “Models of Neocortical Layer 5b Pyramidal Cells Capturing a Wide Range of Dendritic and Perisomatic Active Properties.” Edited by Lyle J. Graham. PLoS Computational Biology 7 (7): e1002107. doi: https://doi.org/10.1371/journal.pcbi.1002107.