GBI-LNMA 2024 course: Image analysis and data reuse through image repositories
https://globalbioimaging.org/international-training-courses/gbi-lnma-2024-course
9:00 - 9:30 Welcome by the director of the Institute of Biotechnology - Laura Palomares (National Autonomous University of Mexico)
9:30 - 11:00 Introduction to digital images - Trainer: Rocco D’Antuono (Francis Crick Institute), Helper: Stefania Marcotti (King's College London)
11:30 - 13:00 Introduction to digital images (continued) - Trainer: Rocco D’Antuono (Francis Crick Institute), Helper: Stefania Marcotti (King's College London)
14:00 - 16:30 Using ImageJ/FIJI via a graphical user interface (GUI) - Trainer: Stefania Marcotti (King's College London), Helpers: Rocco D’Antuono (Francis Crick Institute), Christopher Wood
16:30 - 17:00 Participants are split into (thematic) groups (4-5 participants per group) - Participants & instructors of the course
A visit to the Laboratorio Nacional de Microscopía Avanzada (National Laboratory of Advanced Microscopy) to explore its facilities and imaging technologies
9:00 - 10:30 Introduction to ImageJ macro language - Trainer: Stefania Marcotti (King's College London), Helper: Rocco D’Antuono (Francis Crick Institute)
11:00 - 13:00 Introduction ImageJ macro language (continued) - Trainer: Stefania Marcotti (King's College London), Helper: Rocco D’Antuono (Francis Crick Institute)
15:00 - 17:00 Work in thematic groups (4-5 participants per group) - Participants & instructors of the course
A unique performance by a local folkloric dance group
9:00 - 10:30 Introduction to Python programming (Session #1) - Trainer: Sebastian Gonzalez Tirado (European Molecular Biology Laboratory), Helpers: Rocco D’Antuono (Francis Crick Institute), Stefania Marcotti (King's College London), Adan Guerrero, (National Autonomous University of Mexico)
11:00 - 13:00 Introduction to Python programming (Session #1 continued) - Trainer: Sebastian Gonzalez Tirado (European Molecular Biology Laboratory), Helpers: Rocco D’Antuono (Francis Crick Institute), Stefania Marcotti (King's College London), Adan Guerrero (National Autonomous University of Mexico)
14:00 - 15:00 Managing your image data for the long term: FAIR data, REMBI metadata and the BioImage Archive - Speaker: Matthew Hartley (European Bioinformatics Institute)
15:00 - 17:00 Work in thematic groups (4-5 participants per group) - Participants & instructors of the course
17:00 - 17:45 Venom House Tour - Tour of the facility that includes venomous reptiles (snakes & lizards) that are used in antivenom research and production
9:00 - 10:30 Introduction to Python programming (Session #2) - Trainer: Sebastian Gonzalez Tirado (European Molecular Biology Laboratory), Helpers: Rocco D’Antuono (Francis Crick Institute), Stefania Marcotti (King's College London), Adan Guerrero (National Autonomous University of Mexico)
11:00 - 13:00 Introduction to Python programming (Session #2 continued) - Trainer: Sebastian Gonzalez Tirado (European Molecular Biology Laboratory), Helpers: Rocco D’Antuono (Francis Crick Institute), Stefania Marcotti (King's College London), Adan Guerrero (National Autonomous University of Mexico)
14:00 - 17:00 BioImage Archive image data repository workshop - Trainer: Aybuke Kupcu Yoldas (European Bioinformatics Institute)
9:00 - 10:30 Napari viewer - Trainer: Rocco D’Antuono (Francis Crick Institute), Helpers: Adan Guerrero (National Autonomous University of Mexico)
11:00 - 13:00 Napari viewer & plugins - Trainer: Rocco D’Antuono (Francis Crick Institute), Helpers: Adan Guerrero (National Autonomous University of Mexico)
14:00 - 15:00 Napari superres & zelda plugins - Trainers: Rocco D’Antuono (Francis Crick Institute) & Julian Mejia Morales (National Autonomous University of Mexico)
15:00 - 15:30 Building reusable bioimage analysis workflows with Nextflow - Speaker: Sebastian Gonzalez Tirado (European Molecular Biology Laboratory)
15:30 - 17:00 Presentations from thematic groups (4-5 participants per group) - Participants of the course
17:30 Closing Dinner - Dinner featuring local cuisine and final opportunity to network with participants and the faculty
-
Download FIJI from here.
-
To avoid any permissions issues, install FIJI is in your home directory:
- PC:
C:\users\<your user name> - Mac:
/Users/<your user name>
WARNING: FIJI must be installed in a location where it has write permission - otherwise, it cannot update itself
- PC:
-
Start FIJI and allow the updater to run:
-
(Optional) If the updater does not run automatically, select
Help > Update: -
If FIJI produces any error messages, it is most likely because it does not have the necessary permissions to update itself - return to step #2 and double-check the location of the installation.
- Download Anaconda Navigator from https://www.anaconda.com/download
use Anaconda Prompt or other terminal to create a conda environment (make sure you have no other existing "mynapari-env")
conda create -y -n mynapari-env python=3.8
conda activate mynapari-env
conda install napari pyqt
conda create -y -n np-superres -c conda-forge python=3.9
conda activate np-superres
conda install -c conda-forge napari pyqt ipywidgets git
use graphical installation
or get the latest developments with
pip install git+https://github.com/RoccoDAnt/napari-superres.git
More info at https://github.com/RoccoDAnt/napari-superres
conda create -y -n np-zelda python=3.8
conda activate napari-env
conda install napari pyqt git
use graphical installation
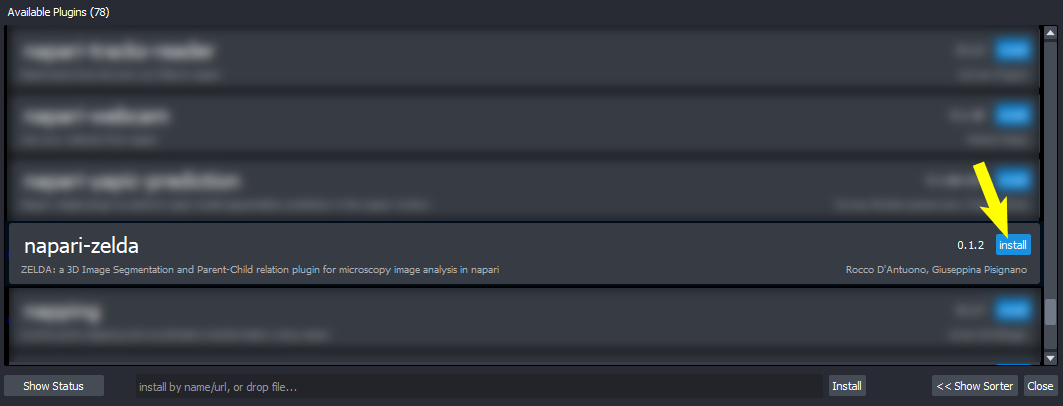
- Plugins / Install/Uninstall Package(s)
or get the latest developments with
pip install git+https://github.com/RoccoDAnt/napari-zelda.git
More info at https://github.com/RoccoDAnt/napari-zelda